Plasmodium knowlesi was initially identified in the 30s as a natural Plasmodium of Macaca fascicularis monkey also capable of experimentally infecting humans. It gained a relative notoriety in the mid-30s as an alternative to Plasmodium vivax in the treatment of the general paralysis of the insane (neurosyphilis). In 1965 the first natural human infection was described in a US military surveyor coming back from the Pahang jungle of the Malaysian peninsula. P. knowlesi was again brought to the attention of the medical community when in 2004, Balbir Singh and his co-workers reported that about 58% of malaria cases observed in the Kapit district of the Malaysian Borneo were actually caused by P. knowlesi . In the following years several reports showed that P. knowlesi is much more widespread than initially thought with cases reported across Southeast Asia. This infection should also be considered in the differential diagnosis of any febrile travellers coming back from a recent travel to forested areas of Southeast Asia. P. knowlesi can cause severe malaria with a rate of 6–9% and with a case fatality rate of 3%. Respiratory distress, acute renal failure, shock and hyperbilirubinemia are the most frequently observed complications of severe P. knowlesi malaria. Chloroquine is considered the treatment of choice of uncomplicated malaria caused by P. knowlesi
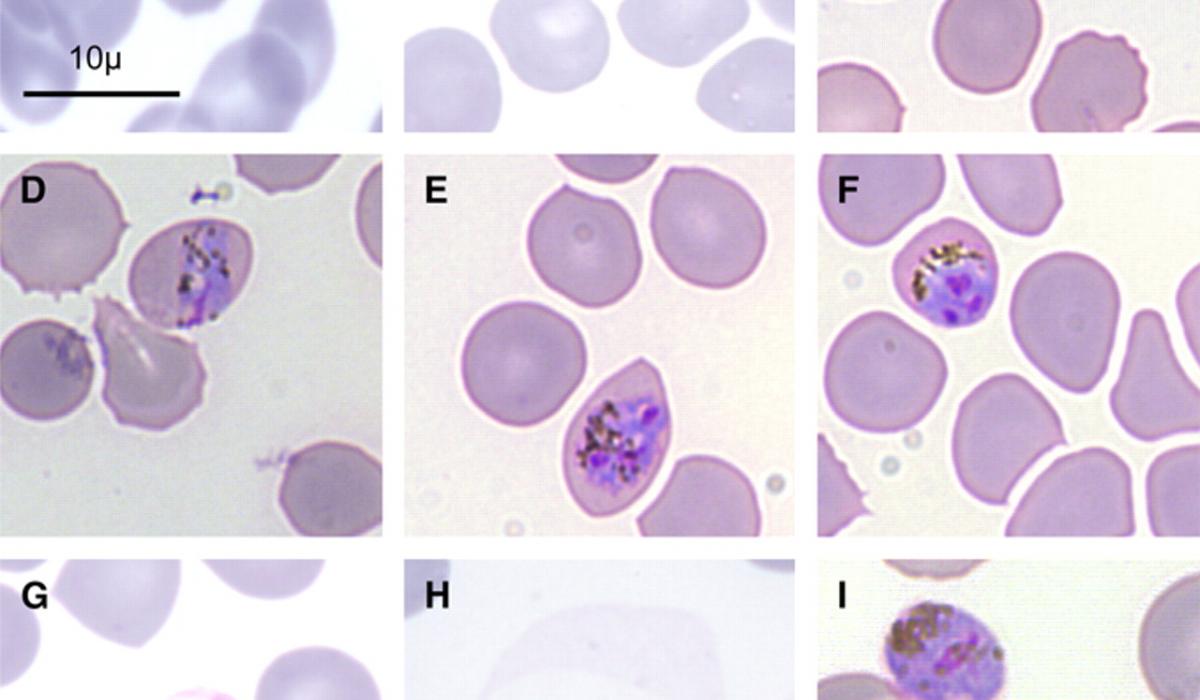
P. knowlesi, considered the fifth human malaria parasite (although a human-to-human transmission cycle has not yet been conclusively documented) shares a close phylogenetic relationship with Plasmodium vivax ( White, 2008 ) and morphological features ( Fig. 1 ) that resemble those of either Plasmodium falciparum and Plasmodium malariae ( Lee et al., 2009a,b ). It differs from P. vivax because of the absence of a latent liver stage (hypnozoite) and the length of its asexual cycle. P. knowlesi is the only primate malaria species characterized by a quotidian (24 h) asexual blood stage development. Similar to P. vivax , P. knowlesi uses the Duffy blood group antigen as a receptor to invade human erythrocytes ( Singh et al., 2003 ). Although the paradigm that erythrocytes of Duffy neg- ative individuals are always refractory to P. vivax infection has been recently challenged by reports from Africa and the Brazilian Ama- zon ( Ryan et al., 2006; Cavasini et al., 2007 ) it is presently unknown if P. knowlesi might also exploit alternative invasion mechanism. P. knowlesi Duffy-binding proteins (PkDBP) localized in micronemes bind on erythrocytes during invasion ( Adams et al., 1990 ); subse- quent invasion of human red blood cells requires the interaction of PkDBP with the Duffy antigen receptor for chemokines (DARC) and after apical reorientation a junction ensues between target eryth- rocytes and invading merozoites. The P. knowlesi Duffy-binding domain contains 12 conserved cysteine residues and its architec- ture is probably constituted by two sub-domains, the major of which contains the C5–C8 region that has been identified as essen- tial for the interaction with the erythrocytes. It has been previously shown that P. knowlesi merozoites are able to interact with Duffy- negative human erythrocytes and that apical orientation take place normally but invasion is aborted since a junction does not develop ( Miller et al., 1975 ). Deletion of gene of P. knowlesi which encodes PkDBP, results in the complete inability to invade Duffy-positive human erythrocytes ( Singh et al., 2003 ). Using the Bayesian coa- lescent approach to estimate the time to the most recent common ancestor (TMRCA) for P. knowlesi , Lee and co-workers estimated the TMRCA for P. knowlesi to be 98,000–478,000 years ( Lee et al., 2011 )
The genome of P. knowlesi is made of 14 chromosomes contain- ing a total of 5188 protein-encoding genes, 80% of which, could be identified in both P. falciparum and P. vivax . The two major variant gene families within P. knowlesi ( i.e ., SICA var and Kir ) are randomly located across all the 14 chromosomes a finding that is in contrast with other Plasmodium genomes ( Pain et al., 2008 ). However, the most striking feature is the molecular mimicry of Kir proteins with near one-half of the CD99 host extracellular domain that is involved as an immunoregulatory molecule in the T-cell response. However, the higher mimicry with host cell receptor CD99 in macaques sup- ports the notion that P. knowlesi is particularly adapted to macaque hosts. The SICA antigens encoded by the parasite’s SICA var gene family, are associated with parasite virulence and were identified at the surface of infected erythrocytes
Initial studies conducted in Malaysia identified Anopheles hack- eri as the vector of P. knowlesi but since this mosquito is not attracted to humans, feeds mainly on monkeys and is found only in the deep jungle, it was stated that P. knowlesi malaria would not be a serious problem for humans ( Wharton and Eyles, 1961; Chin et al., 1968 ). After the recognition of the importance of P. knowlesi malaria in Sarawak (Malaysian Borneo), vector studies conducted in three different ecological niches ( i.e ., forest, farm and villages) in the Kapit district identified A. latens, which belongs to the Leucos- phyrus mosquitoes group, as the main natural vector of P. knowlesi for both monkeys and humans ( Vythilingam et al., 2006; Tan et al., 2008; Vythilingam, 2010 ). The geographic distribution of the Anopheles leucosphirus group has been shown to range from South-western India, eastward to Southern China, Taiwan, mainland Southeast Asia, Indonesia and Philippines ( Sallum et al., 2005 ). Experimental laboratory studies carried out to verify sus- ceptibility to P. knowlesi of Anopheles freeborni, Anopheles quadrimaculatus, Anopheles maculatus and Anopheles balabacensis showed that A. quadrimaculatus was naturally resistant to the par- asite while A. balabacensis emerged as the most efficient vector producing more than 1000 sporozoites and inducing infection in monkeys after a prepatent period of 7–8 days ( Vythilingam et al., 2008 ). Another study conducted by the same group in the state of Pahang revealed Anopheles cracens , a more anthropophilic member of the Leucosphyrus group, as the vector of P. knowlesi (as well as of P. cynomolgi ) in that area ( Vythilingam et al., 2008 ). In the Sabah region, A. balabacensis has been incriminated as a possible vector of P. knowlesi ( Vythilingam, 2010 ). Finally, in Vietnam, Anopheles dirus , a member of the Anopheles leucosphyrus group has been impli- cated as the vector of P. knowlesi infection ( Nakazawa et al., 2009 ). A survey conducted in the forest and forest fringe areas of Khanh Phu (Southern Vietnam) from 2008 to 2010 showed that A. dirus mosquitoes are the only malaria vectors in that area and near 1.6%are infected with sporozoites. Using PCR analysis to distinguish malaria species, it was shown that 43% of salivary glands assayed were PCR positive for P. knowlesi with frequent coinfections (71% of examined glands) and with the combination P. knowlesi / P. vivax as the most common ( Marchand et al., 2011 ). It is worth noting that in Singapore, where several autochthonous cases of human P. knowlesi malaria were reg- istered, mosquitoes belonging to the A. leucosphyrus group were not found. It has been suggested that in Singapore, Anopheles kochi might be the potential vector of P. knowlesi and the fact that is a highly zoophagic mosquito could explain the small number of cases of knowlesi malaria despite the high frequency of deployed military personnel in the forested areas ( Jeslyn et al., 2011 ).
| BACK | Source: ELSEVIER Journal |
