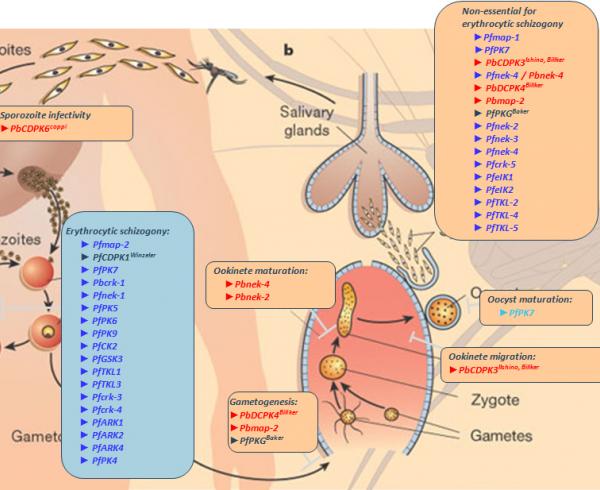
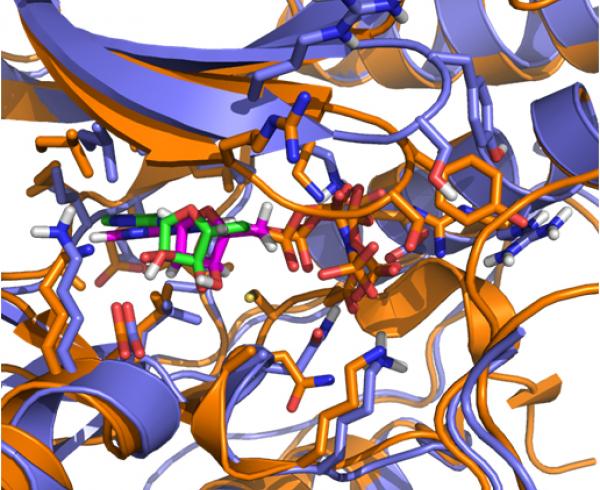
Introduction
Proteins are used by cells as tools to carry out most of their biological processes, ranging from the respiration or metabolism of nutrients, to the sending and processing of intra- and intercellular signals, and to maintaining cellular shape, ensuring growth and controlling cell division. The activities of the proteins responsible for these activities can usually be regulated by binding small molecules to them, a property that is often used in rational drug design.