
Within the FP7 SYNERGY project Douglas Connect initiated two collaborative research project pilots in 2010: one in Drug Design and one in Predictive Toxicology. The work is being carried out by a range of individual organisations from industry and academia located in several countries ie. they formed a Virtual Organisation (VO). Services supporting the collaborative work were tested during the pilots.
Scientists Against Malaria (SAM) was formed by Douglas Connect from the InnovationWell Neglected Diseases Collaboration Pool as a virtual drug discovery organization to collaborate on the design of kinase inhibitors against the Plasmodium falciparum malarial parasite.
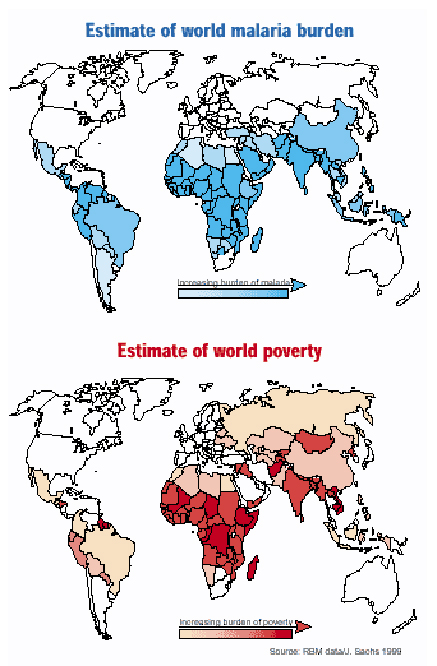 The most severe form of Malaria is caused by the parasite Plasmodium falciparum. Malaria causes 1-3 million deaths every year, the vast majority being children under 5 years old. Poverty is both an effect and cause of Malaria. The malaria burden hinders economic development; the poor often cannot afford measures to prevent or treat the disease.
The most severe form of Malaria is caused by the parasite Plasmodium falciparum. Malaria causes 1-3 million deaths every year, the vast majority being children under 5 years old. Poverty is both an effect and cause of Malaria. The malaria burden hinders economic development; the poor often cannot afford measures to prevent or treat the disease.
With the goal of developing new inhibitors active against the parasite and the potential subsequent development of Malaria Treatment: Lack of Investment a new therapeutic against malaria, we decided to use the Plasmodium falciparum mitogen-activated kinase 2 (PfMAP-2). The protein is essential for parasite survival but not related to any human kinase, thus offering the potential to find parasite-selective inhibitors.
The SAM research activity required a number of roles to be filled by partners. Christian Doerig provided knowledge on parasite biology and target selection, Roman Affentranger created homology models of initial and final target proteins, the University of Cincinnati Drug Discovery Center (UC DDC) provided their compound library, Alessandro Contini, Hugo Gutierrez de Teran and Matt Clark were responsible for computational aspects of the project, Monash University produced protein for the assays, UC DDC developed, optimized and carried out the experimental assays.
Toxicity predictions are planned to flow in to SAM from the predictive toxicology pilot project which is being run in parallel within the OpenTox framework for predictive toxicology.
To support the pilot activity, we designed a combination of interoperable information systems, ontologies and web services. Key collaborative lab notebook infrastructure was provided by Rescentris using their Collaborative Electronic Research Framework (CERF) to manage the data, documents, computational and assay results. Services were designed for integrating activity, toxicology and consensus predictions into a dashboard to track project progress and to support decision making. CERF was integrated with the SYNERGY services CPA (Collaboration Pattern Assistant), CEPS (Complex Events Processing System), and CMS (Collaboration Moderator System).
SAM WorkflowData produced within SAM as well as protocols for computations and experiments are documented in CERF. The system facilitates the import of and display of data and communicates new results in real-time to the Petals enterprise service bus through ontology-based messages. Several additional SYNERGY services subscribe to Petals and receive the data messages. These services support consensus scoring, detect complex event patterns in the data and suggest collaboration patterns based on these complex events patterns.
The >300’000 compounds of the UC DDC library were expanded to >1.3 million molecular structures (including protonation, tautomeric and isomeric states). These structures were screened independently using three different docking tools (AutoDock, Vina, Glide). The three scores were combined using a consensus scoring strategy, based on which we selected 996 compounds for experimental assays. Assays for these compounds are currently in progress. In parallel, a pharmacophore-based screen in combination with a free-energy calculation predicted ~700 potential inhibitors, which were assayed by UC DDC. Initial results look promising (low-micromolar inhibitors), and dose-response curves are currently being determined.
